Deep within our cells, proteins run along precisely scheduled molecular pathways to deliver their chemical messages. Scientists are looking for ways to slow these reactions down, speed them up, or turn them off entirely in the hopes for new treatments for illnesses such as Parkinson’s disease and cancer.
Certain protein messengers, known as G-alpha proteins, show up in a wide range of organisms. Scientists have found them in humans, nematodes, and even plants. These proteins play a vital role in basic cellular division processes, and they also act as messengers between us and our environment. G-alpha proteins help us to see, taste, reproduce. Yet it took years of investigation before anyone knew how G-alpha proteins kept time.
Almost a decade ago, while working at a pharmaceutical development company, David Siderovski discovered a new class of protein regulators that conduct our body’s G-alpha symphony. These regulators are known as RGS proteins (regulators of G protein signaling). Since then, scientists have been investigating the potential of RGS proteins to alter G-alpha function and control the timing of hormone and neurotransmitter signaling. In patients with Parkinson’s disease, for example, scientists identified a set of neurons in the brain that die off, leading to a loss of the neurotransmitter dopamine — effectively inhibiting a critical neurological pathway. This loss of dopamine signaling is commonly treated with L-dopa, which the brain converts into dopamine. “So what they’re trying to do is push the gas pedal with dopamine,” Siderovski says. “But another way to get the car lurching down the road is to snip the brake lines.”
With such a well-known drug-targeting potential for G-alpha proteins, Siderovski’s discovery of a new group of their regulators opened up several promising paths of investigation for drug development. But he found that time was an issue. His company wanted him to turn a profit on the new RGS proteins at a rate that wouldn’t allow him to explore their fundamental principles. “They put the screws to me and said, ‘Show us the clear shareholder opportunity in these proteins’,” he says. “And I needed the time to flesh them out.” After all, there wasn’t only one RGS protein; there were forty. Only a few were going to be really viable drug-discovery targets, and the rest a waste of time — at least according to corporate sentiment.
With forty RGS proteins to investigate, Siderovski tried to narrow down the search. Where were RGS proteins expressed in our bodies? Were any of them involved with a pathological condition? How could he know that one protein, say RGS7, is a better target than RGS9 to help people with the movement disorder associated with Parkinson’s disease?
It was time to get back to the basics. In order to decide which of the RGS proteins to pursue, Siderovski needed to understand the mechanics of cell messaging in greater depth. When hormones or neurotransmitters send messages to proteins inside a cell, receptors embedded in the cell’s membrane “talk” to the complicated machinery on the inside. This kicks off the cell’s interior delivery system and creates a second tier of messengers. The second tier may consist entirely of a single wandering ion, but often it’s a whole cascade of reactions that eventually provokes the intended cellular response.
But how does the G-alpha protein talk to its receptor? And how can RGS proteins alter the timing of the process? Siderovski knew that the receptor’s structure would reveal some of the answers. The receptor winds gracefully in and out of the cell’s membrane seven times before ending in the G protein, a three-part set of subunits inside the cell. When the receptor stimulates these subunits from outside the cell, they detach and re-bind to one another.
From his industry-based lab, Siderovski considered these issues. He wanted to see it all happening. He would have to get a snapshot of the receptor and its subunits in action. But because of the receptor’s complicated structure, no one had ever taken that picture.
An easy choice
Such an investigation would not have been shareholder-friendly in the short term, Siderovski says. “We don’t have the luxury of time in industry. The shareholders want the pipeline filled quickly with viable drugs that are going to get FDA approval and be billion-dollar sellers.”
So he moved to UNC, and it was only then that he realized just how much industry had been doing for him. “When something breaks, I have to run in with my gloves and repair the hose that’s leaking CO2,” he says. In addition to fixing equipment, he hires his research staff, orders supplies, writes grants, and sells the “product” at scientific conferences. “My analogy to my kids is that I’m running a diner and the kitchen has always got a grease fire, because that’s the way I feel. All those things that you have in industry done for you as part of ‘the team,’ you basically do yourself here.”
Besides creating his own timeline for investigation, Siderovski gets to collaborate with a group of similarly passionate folks in what he calls the “academic G protein signaling scene,” which allows him to use a broad range of tools to study the proteins that he loves. Siderovski’s UNC lab has used everything from mouse genetics to X-ray crystallography, allowing him to better understand how G-alpha proteins and their regulators work together.
“I’m a master generalist, and my dad was like that too,” he says. “My dad was an electrician. But he did plumbing, and he did construction and maintenance and grouting and mortar and brick. And I do that scientifically.” Siderovski’s lab, for example, has published mouse genetic studies. The team isolated the gene for a particular RGS protein, knocked it out of the mouse’s DNA, and explored how the loss of that RGS protein expresses itself in the animal’s behavior and in its responses to hormones. But the most helpful tool he’s adopted involves bombarding proteins with X rays in order to create mathematically derived “photographs” of their molecular structures.
This technique is called X-ray crystallography, and he emphasizes that he’s not actually trained to do it. “I know a little bit about how to get crystallography done,” Siderovski says. “I know a little bit about how to make mouse models, and so through that spectrum I’m trying to learn about these hormone receptors and how they’re acting and how they’re shut off.”
Armed with X rays, Siderovski’s lab can finally observe the proteins, understand how they work, and manipulate the rate at which signals move from one end of the receptor to another. But the receptors in their natural form aren’t very photogenic. Before graduate student Christopher Johnston could snap the latest molecular picture, he had to figure out a way to make the proteins organize themselves into an orderly crystal lattice. X rays aimed at that lattice will diffract into a pattern that can then be mathematically reconstituted into the snake-like shape of the atoms in the original protein. And once again, Siderovski and his research team found the help they needed in what he calls the “beautiful and workable infrastructure” of the UNC protein scene.
“There’s a number of people on this campus who are world experts in this technology,” he says. “And the joy of Chapel Hill is that we all share and collaborate.” So Siderovski’s lab was able — with the help of fellow pharmacologist John Sondek — to catch the receptor in the action of binding, activating, and talking to its cellular partner — the G-alpha protein.
Siderovski gets his snapshot
His lab did it without the pressure of working under what he calls industry’s “biomedical relevance mandate,” and now Siderovski has created his own definition of that term. “Everything that we do is biomedically relevant,” he says. “But with what timing? Is it next week we should go to the compound library and screen for the inhibitor? Or do we need another five years?” He wants to drive RGS proteins toward the development of a drug that eases suffering or saves lives.
Contrast this with the industry’s definition of biomedical relevance, which Siderovski now describes more precisely as “direct drug discovery applicability.” It’s a relevance to the market, but not necessarily to medicine. It’s the ability to predict a blockbuster pill before you’ve designed the investigations into the compounds the pill will contain.
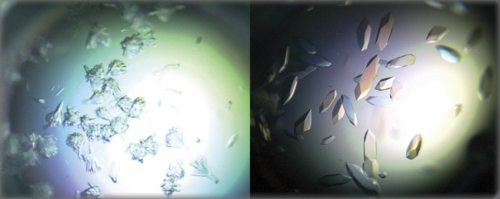
In the end, semantics matter less than the evidence. Siderovski opens a file on his laptop and scrolls down to a series of photos embedded in his most recent grant proposal. One of the photos is the first-ever crystal snapshot of a G-alpha protein caught in activation. It’s a picture that scientists around the world have spent a lot of money trying to capture. “So this is kind of a before-and-after shot,” he says. “In the first one these are really nasty-looking crystals. They’re not going to shoot in an X-ray beam very well. But then we added a little peptide to stabilize the protein, and now look how beautiful they are. Isn’t that nice?”
David Siderovski is an associate professor in the Department of Pharmacology at UNC. He won the Phillip and Ruth Hettleman Prize for Artistic and Scholarly Achievements by Young Faculty in 2006. John Sondek is a professor of biochemistry at UNC.