In the United States alone, 60 to 70 million people suffer from digestive disorders like Crohn’s disease, ulcerative colitis, and irritable bowel syndrome, according to the National Institutes of Health. These diseases range in severity, from minor symptoms that lead to missed school or work to chronic and debilitating illnesses requiring continuous medical care. In fact, more male employees miss work due to digestive conditions than any other reason.
The entire lining of the gut is approximately 300 to 400 square feet — the size of a small studio apartment. And its tenants include trillions of microorganisms responsible for building immunity, processing toxins, and releasing 95 percent of the body’s serotonin, which relieves anxiety and depression. It’s like the upstairs neighbor that’s constantly partying; you can hear and even feel what’s happening, but can only imagine what’s actually taking place. This is why disease of the digestive tract can be hard to diagnose.
There’s just a lot of cells and tissue. So understanding wherein the problem lies can be tricky.
But what if your gastroenterologist could, through a minor procedure, take a small amount of your own intestinal cells and test different treatments against them to uncover what could cure, or at least lessen, your symptoms?
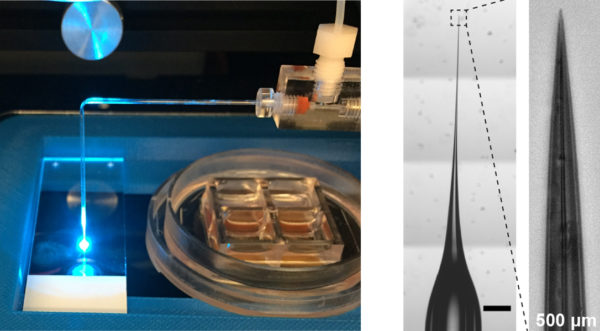
Scott Magness, a UNC geneticist, cell biologist, and biomedical engineer, has done exactly that. He and Nancy Allbritton, chair of the UNC/NCSU Joint Department of Biomedical Engineering, have created a “gut on a chip” — a small-scale version of someone’s gut lining on a surface area the size of a postage stamp. This research has been so successful that they’ve created a spinoff company, called Altis BioSystems, from it.
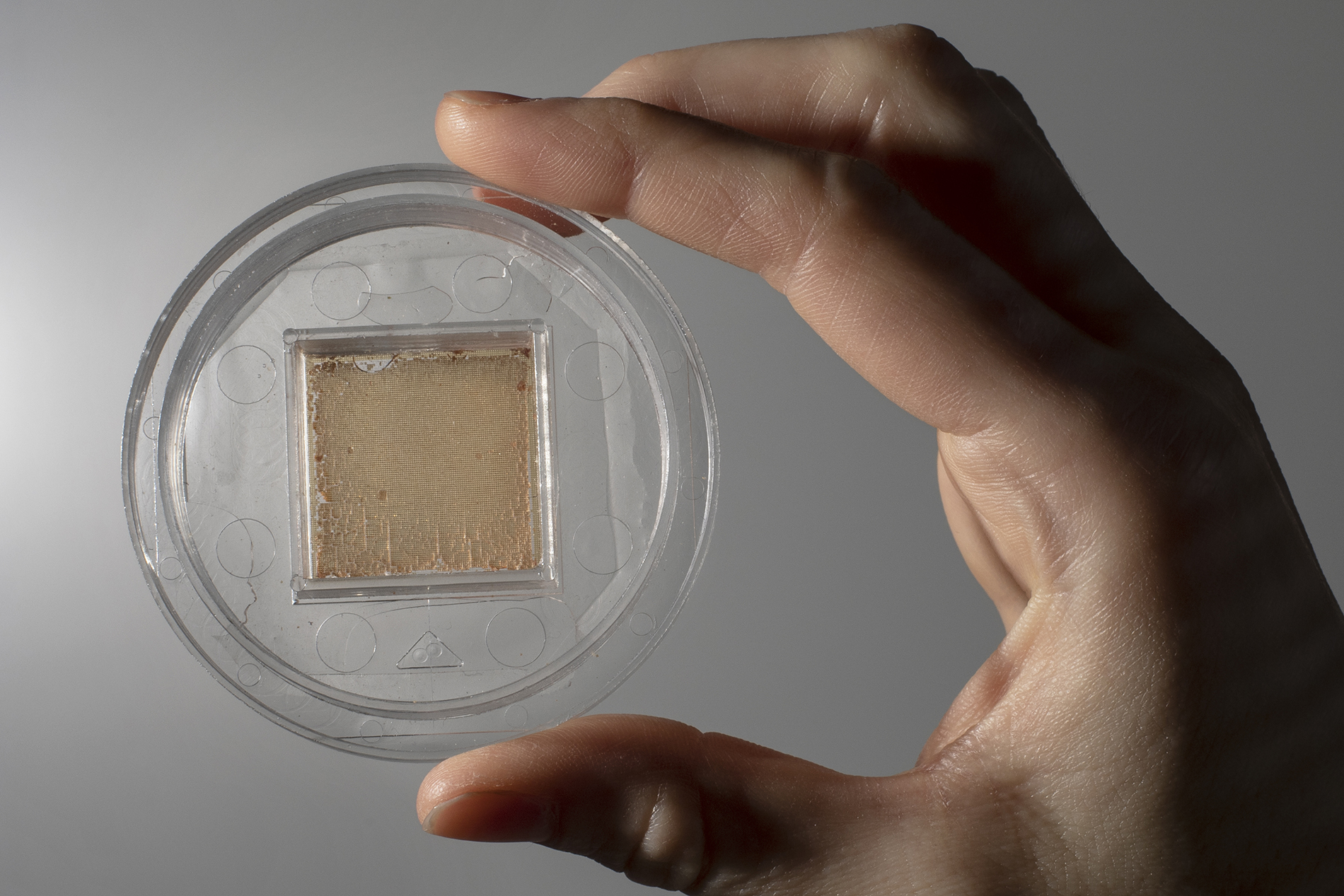
Allbritton’s gut-on-a-chip can culture 15,000 stem cells at once. Look closely at her device and you’ll see its tiny, individual wells.
Basic science research like this, though, is rarely so immediately applicable. How did they do it? Patience, hard work, and the creation of novel tools.
Solving the stem cell mystery
Since the early 2000s, Magness has been captivated by tissue regeneration. During that time, while working on his first postdoc at UNC, he was studying a rare disorder called protoporphyria — an enzyme deficiency that can lead to severe liver disease. He’d run these experiments where he would cut a liver nearly in half and watch it regenerate in just two weeks. “That fascinated me,” he says. “How a whole organ can have the information in its cells to grow back.”
Then, for his second postdoc, he spent three years studying the genes that control tissue-specific stem cell behavior in the retina and spinal cord. Tissue-specific stem cells are specialized in that they only give rise to more cells of the same tissue, while embryonic stem cells become all the different cell types of the body like red blood cells, brain cells, and muscle cells.
Most people are familiar with embryonic stem cells, which garnered public attention in the 1970s when abortions first became legal, opening the ability to study embryos. At that time, a ban was quickly placed on funding for embryonic stem cell research. But in 1995, the ban funding for the study of stem cells left over from in-vitro fertilization, specifically, was lifted.
Tissue-specific stem cells, on the other hand, have been in use since the 1950s, when bone marrow transplants were first used to treat leukemia. They are found throughout the body after development and multiply by cell division to replenish dying cells and regenerate damaged tissues.
Magness studies adult stem cells, specifically intestinal and stomach stem cells. After completing his second postdoc in 2006, he applied for a job in the Division of Gastroenterology and Hepatology at Carolina. Prior to starting his faculty position, he had never considered studying the lining of the gut. Because it’s the most regenerative tissue in the body, he assumed someone must have figured out its stem cell biology long ago.
“But it turns out that wasn’t the case,” Magness says. “There were no markers for intestinal stem cells. You couldn’t identify them, you couldn’t isolate them, you couldn’t grow them. And that’s a very daunting challenge — to study something you can’t find.” But that only intrigued Magness. Challenge accepted.
Magness and his team began investigating a gene called Sox9, which, they discovered, is expressed in intestinal stem cells and could be used to identify them. Once identified, and isolated, growing them in a petri dish quickly became their next order of business. To do this, they would have to “trick” the stem cells into thinking they were still within the body by recreating the complex nature of the intestinal stem cell environment.
At the same time, Dutch geneticist Hans Clevers and his group were attempting that exact feat. “We were working in parallel, unknowingly,” Magness points out. “We had only one or two missing factors.” Clevers beat him to the punch, publishing his results in Nature. But Magness’ lab quickly followed suit and, in 2010, Magness and graduate student Adam Gracz became the first researchers in the United States to grow intestinal stem cells outside of the body.
While this development marked a huge success for the lab, they soon hit another roadblock. After isolating cells using stem cell markers, they discovered that only one out of every 100 could actually self-renew like stem cells are supposed to.
Previously, scientists believed stem cells were a distinct population within the body, but the definition is much broader than that. Turns out, many cell populations have stem cell properties — but only under certain conditions or environments. “There are cells who don’t know who they are yet in life,” Magness says. “I call them ‘teenager cells’ because they’re not quite adults, but they’re not quite babies either. And they are very flexible in how they can adapt to the environment.”
The intestine contains both “active” stem cells and “reserve” stem cells. If the active cells get depleted, the body sends a signal to the reserves, notifying them to “wake up” and replenish the cells that were damaged. “And it’s very difficult to identify the reserve ones,” Magness says. “They’re rare, hiding and not proliferating rapidly. There’s no growth. So they are extremely hard to find and study.” Magness and his team went in search for better tools to study what affects their growth and how to wake them up.
Growing mini-guts, on a chip
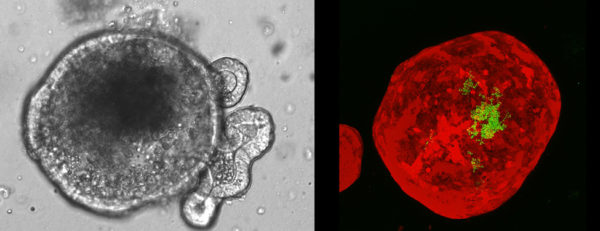
Left: A microscopic view of a mini-gut. Right: A mini-gut (red) that has had bacteria (green) injected into its hollow center.
In 2012, Magness began collaborating with Nancy Allbritton on this problem. A UNC biomedical engineer, Allbritton specializes in microfabrication, which involves packing manufacturing processes into small devices like the postage stamp-sized structure called a “cell-raft array” she created to culture 15,000 cells at once — the first tool in a series of powerful bioengineered devices and platforms that mimic the gut lining on a chip.
Allbritton’s invention contains thousands of microscopic wells arranged in a grid, each well as wide as a strand of hair, for studying individual cells. This allowed Magness to observe thousands of single stem cells at once and discover that an intestinal nurse cell, a cell that provides stability for neighboring cells, must touch the stem cell to help it survive and grow into a spherical structure with a hollow center — something Magness calls a “mini-gut.”
“So we can take a very small piece of tissue — like the size of a couple grains of rice — from the intestine and grow the gut lining as mini-guts in a dish,” Magness says. “Just imagine what we can do with that, the studies we can do.”
Magness believed he could use these mini-guts, called organoids, to study the various species of bacteria that live on the gut lining. Most are “healthy” strains, but some contribute to problems like diarrhea and inflammatory bowel diseases. Gut bacteria are hard to study in the lab, though, as they are highly sensitive to oxygen and die when exposed to air.
Ian Williamson, a graduate student in Magness’s lab, built a microscopic device similar to a sewing machine that could inject bacteria through a needle into the center of mini-guts cultured on the chip — and it could do this very fast, in hundreds of mini-guts in one experiment. With this technology, Magness’s group could protect bacteria and study their effects on the lining of the gut.
Mimicking gut architecture
Unlike the intestinal cells inside humans, mini-guts grow into small balls and lack the same structure – or architecture – as the gut lining. To address this challenge, Magness and Allbritton teamed up with UNC computational biologist Shawn Gomez and geneticist Scott Bultman to create a fully functioning lining of the human gut that fits on a chip the size of a postage stamp. The technology uses human cells taken from biopsies and transforms them into the complex architecture of the gastrointestinal tract, rather than a disorganized ball of cells.
The intricacies of this organ system stem not only from its size — it’s 30 feet in length — but the millions of pockets packed with different types of regenerative stem cells. The gut-on-a-chip tackles these complexities by combining a polymer base with a series of sponge-like molecules, called hydrogels, which provide the structural support and biochemical cues for growing cells from the intestinal lining. Plumbing attached to the device then sends chemicals, fluids, and gases inside of it to instruct the cells on how and where to grow.
This group is not the first to put an organ on a chip. Other researchers have developed similar platforms as they reduce the cost and inaccuracies of animal models and speed up treatment times compared to traditional drug discovery, which can take decades and billions of dollars to complete. This technology, though, is some of the first to use patient-derived tissues and recreate the in-depth functions of the gut.
From this work, Magness and Allbritton co-founded Altis Biosystems, a biotechnology company that develops and manufactures research tools to accelerate drug development. “A lot of drugs fail because they act differently on you than they do me,” Magness explains. “And we don’t know that until you try the drug. So there’s been a lot of interest in having better, more predictive models about how drugs will behave with a specific person. And what we’ve created is a platform for this kind of personalized medicine.”
Testing drugs, one disease at a time
Altis Biosystems’ technology can be used to test the toxicity of chemotherapeutic compounds, better understand how probiotics work, and improve intestinal barrier problems — known generally as leaky gut — which occur in conjunction with several diseases such as rheumatoid arthritis, diabetes, and even Alzheimer’s disease. People in the livestock and poultry industries are also interested in utilizing this novel tool, as animal diet and digestive health plays a large role in their mortality rates.
“This is a really powerful, transformative technology,” Magness says. “I think it’s important to point out that this is the first time we can study normal intestinal tissue. Drug companies have been using colon cancer cell lines to study all of these processes, and many times they failed to predict drug efficacy in clinical trials, causing the loss of millions and millions of dollars.”
The next step of this research involves merging multiple tissue types found within the gut onto a chip. “Underneath the gut lining, it’s kind of like the Grand Canyon,” he says. “You have all these different layers of cells.” Four layers of specialized tissue sit below the gastrointestinal lining.
Right now, Magness is working with NC State engineer Michael Daniele to deconstruct the layer containing blood vessels and put that on a chip to study how the gut lining and its vessels communicate. They’re doing this to learn more about hypoxic injury — when there’s not enough blood flow to the intestines, which cuts off the oxygen supply.
“It’s basically a stroke in the gut,” Magness says. “And if that happens, the gut lining will die very quickly.” Magness will use this device to learn more about how stem cells function in low-oxygen environments and its ability to renew blood vessels for transplantation into the body.
By more closely observing stem cells in the other layers of the gut, Magness hopes to uncover new information about complex diseases.
“It’s super exciting — and very creative,” he says. “It’s basically taking biology that happens every day and trying to recreate that super complex biology that’s evolved over millions of years in a dish so that it works that same way it does in the body. That challenge, the puzzle of doing that, is super satisfying and motivating. It’s like making little mini-works of art.”