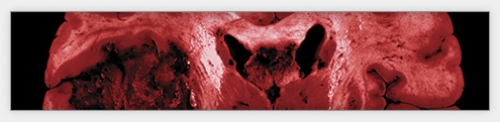
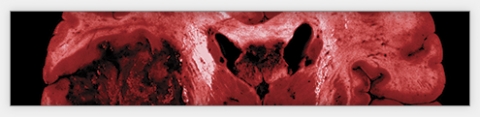
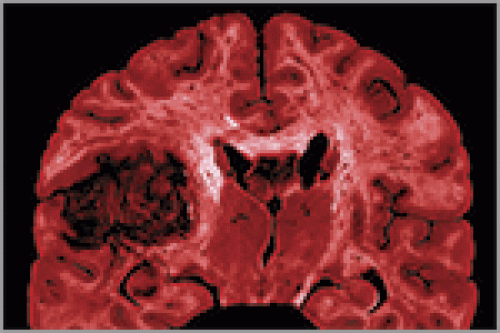
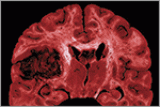
As part of the Cancer Genome Atlas network, oncologists Neil Hayes and Charles Perou study changes in gene activity that could spur cancer. Last September, Hayes and a team of scientists found that the activity of a handful of genes was unusual in the brain cells of patients suffering from a kind of brain tumor called glioblastoma.
Scientists now think of cancer as a constellation of diseases in which the body loses its ability to put the brakes on cell growth and movement. The changes that cause the brakes to fail may be situated anywhere in the genome. Personalized medicine, which tailors therapy to the individual’s genetic makeup, could revolutionize how doctors treat cancer. But to truly personalize treatment, oncologists need to understand all the genetic abnormalities that could trigger the disease.
Mining the genetic information hidden in cancer cells is no mean feat. There’s a whopping number of nucleotide bases to parse; the human genome contains about twenty-five thousand protein-coding genes, according to the National Human Genome Research Institute. And that doesn’t include the more than 98 percent of our genome that’s made up of noncoding DNA, most of whose functions scientists have yet to plumb.
To confront the challenge, the network brought together eighteen institutions that are trying to improve doctors’ ability to diagnose, treat, and prevent cancer. The scientists are documenting changes in the composition and behavior of DNA from tumor tissue removed during treatment. The tissues are cataloged and stored without any identifying information in a central repository in Phoenix, Arizona. Researchers at one or more of eight participating institutions analyze samples of each cancer tissue. They then make the documented genomic changes available in public databases, which scientists everywhere can mine to figure out which of those changes might have triggered the cancer.
Last September, the network published in the journal Nature the first results of its comprehensive study of glioblastoma, a recurrent, malignant brain tumor that the National Cancer Institute (NCI) estimates killed thirteen thousand people in the United States last year. Most glioblastoma patients die within fourteen months of diagnosis.
The scientists analyzed the genomes of about two hundred glioblastoma patients whose tumor samples had been stored at the repository in Arizona. They then compared the patients’ “cancer genomes” with their normal genomes — prepared from the patients’ unaffected blood cells.
Hayes says the team chose glioblastoma for their first study because it presented the fewest technical challenges. “Scientists at NCI felt that glioblastoma was less complex and easier to profile for mutations and gene expression changes than other cancers,” he says. Unlike other cancers, glioblastoma tumors show little morphological variation among patients, making glioblastoma a relatively easy subject for a pilot study. “That, of course, doesn’t mean it’s easy to treat,” Hayes says.
Hayes says it’s hard to find tumor samples of suitable quality for analysis. “We need at least 200 milligrams of brain tissue from each patient to extract the DNA and the RNA for analysis,” he says. But the scientists had access to sufficient numbers of glioblastoma tumors, which was another reason they chose to study that type of cancer.
The team found several genetic changes in the tumors. At the time the study went to press, the team had complete sequence information for about half of the two hundred samples, and in all of those sequences eight genes were significantly mutated. In addition, an array of genes involved in cell division and in intracellular growth signaling showed altered copy number — the number of copies of a gene present on the chromosome. Some genes had too many copies; others had too few. There was also abnormal activity in the genes involved in growth regulation, powering some biochemical pathways that lead to cancer-cell proliferation while short-circuiting others that lead to cancer-cell death.
Hayes says the experiment is the most comprehensive analysis of glioblastoma tumors to date. Perhaps the team’s most significant contribution, he says, was to present a network view of brain tumors by turning the spotlight onto a mix of players from interconnected as well as far-flung cellular pathways. That illumination is crucial for tailoring therapy and forming prognoses.
One of the biggest challenges facing the team is to identify the exact points where the problem begins in the pathways of cancer cells. “If you knew those points, you could target your anticancer therapy downstream to those points. If you don’t know, you can hit the tumor cell all day long upstream of those points and see no effect,” he says.
Such a pathway-driven approach to treatment has become the cancer community’s M.O. The trend in cancer therapy has steered away from treating tumors based on where they are in the body, and toward targeting them based on their genetic signatures. “That allows us to think about therapies in a rational way,” Hayes says.
In a nod to this approach, the team also uncovered a novel mechanism of resistance to the frontline brain-tumor drug temozolomide. Oncologists prescribe the drug to patients regardless of their genetic makeup — largely for lack of an alternative therapy — but have long known that patients whose tumor cells have a modification in one gene respond better to the drug.
The gene, MGMT, repairs damaged DNA. Its modified form, which involves the addition of a methyl group to the gene, makes patients responsive to temozolomide. That’s because the drug kills tumor cells by damaging their DNA. Tumor cells with a methyl group attached to MGMT are unable to repair DNA damage; methyl groups muffle the genes they’re attached to, producing an effect scientists call gene silencing.
The link between MGMT methylation and response to temozolomide is old news, but the team found that the methylation of the MGMT genes in some glioblastoma patients could also make normal brain cells prone to mutations because of impaired DNA repair. They observed more mutations, and different kinds of mutations, in the brain cells of patients whose MGMT genes were methylated. This information could help oncologists evaluate the best treatment option for such patients should alternatives become available in the future.
“I think we’ve shed a little light on the important targets and their prevalence in glioblastoma tumors,” Hayes says. To oncologists used to shooting in the dark, that little light represents a whole new way of looking at brain tumors.
Prashant Nair is a master’s student in medical journalism at Carolina.
Coauthors of the study were Neil Hayes, an assistant professor of hematology and oncology, Charles Perou, an associate professor of genetics and pathology, and Michael Topal, a professor of pathology, all in the School of Medicine. The National Institutes of Health, which created the Cancer Genome Atlas network, funded the study. The Lineberger Comprehensive Cancer Center is one of eight Cancer Genome Characterization Centers in the network; other members include MIT’s Broad Institute, Harvard Medical School, Memorial Sloan-Kettering Cancer Center, Lawrence Berkeley National Laboratory, the Sidney Kimmel Cancer Center at Johns Hopkins University, HudsonAlpha Institute for Biotechnology, and the University of Southern California’s Epigenome Center.