In a back room of Kevin Weeks’s lab, a section of wall is covered with results from his early HIV experiments. The printouts look like electrocardiogram readings gone wrong: a series of spikes and dips without any rhythm or pattern. But Weeks and his lab took these zigzagging figures and turned them into the first-ever model of an entire HIV-1 genome.
About five years ago, Weeks and his collaborators came up with a technique to map the structure of short pieces of ribonucleic acid (RNA) down to the level of nucleotides, the smallest pieces of a genome. Weeks is a chemist, not a virologist — his main interest was in solving the puzzle of how RNA genomes are put together. But he also decided to put his research to practical use as soon as he could. “We wanted to apply it to a problem that would really matter,” Weeks says.
He set out to decode the genome of HIV, an RNA-based virus. But when he went looking for funding for the project, he found skeptics. Other researchers were structurally mapping only short pieces of RNA; HIV’s genome is more than nine thousand nucleotides long. If he had written five years ago that they wanted to model an entire HIV genome, he says, “it’s fair to say we’d have been laughed out of the study section.” Instead, the lab kept trying to improve their new technique, and by 2006 they’d made enough progress to get National Institutes of Health funding for a crack at the HIV-1 genome.
To see what makes HIV’s genome — or any long piece of RNA — so difficult to figure out, compare it with the better-understood genetic molecule DNA. “We’re all familiar with DNA’s structure — that double-stranded helix,” Weeks says. Not every piece of DNA looks like that, but most do: a twisting ladder made up of orderly base pairs of nucleotides. “So when you know a sequence of nucleotides,” Weeks says, “you usually know the DNA’s structure, too.”
RNA is different. It’s made up of a long chain of nucleotides, but it’s usually single-stranded. Even when — as in the HIV genome — RNA strands come in pairs, they aren’t linked up nucleotide by nucleotide like DNA. Instead, each strand folds to create tiny structures of its own. An RNA strand folds so closely in places that its nucleotides pair up with each other, making little double helices and loops scattered throughout the genome.
Scientists have had a hard time interpreting this structured jumble of genetic code, mostly because the chemical processes they used to map what individual nucleotides were doing didn’t give consistent results. But Weeks’s lab developed a new method for measuring nucleotide flexibility: how likely nucleotides are to form single-stranded loops and curves, or, if they’re less flexible, rigid helices and base pairs.
The technique is based on a simple idea, Weeks says. “It’s in the chemistry we teach our second-year undergraduates.” Each nucleotide of a piece of RNA is treated with an organic compound. If the nucleotide contains chemical bonds that hold it in a rigid formation, it doesn’t react much to the compound. These are the nucleotides that are more likely to pair up with each other. If there are fewer of those bonds, the nucleotide reacts more strongly, showing that it’s free to form a looping structure. The zigzagging lines on Weeks’s wall are measurements of how strongly nucleotides reacted.
“If you know enough about what parts of an RNA are flexible, you can use that information to make hypotheses about how it looks,” Weeks says. To help do this, they run a computer program that translates the reactivity data into complete pictures. Weeks and Joe Watts, a postdoc in the lab, did this with RNA from HIV-1 particles grown specially for them at the National Cancer Institute in Maryland. They saw a genome that was full of loops and double helices that no one had ever identified before.
By Kevin Weeks, Joseph Watts, and Jason Smith
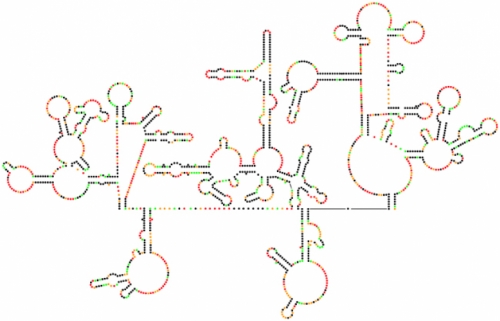
Scientists have sequenced the genetic code of HIV-1 many times, but this map is their first complete look at the genome’s intricate structure. Each dot represents one nucleotide. Black dots are the nucleotides most likely to arrange themselves in base pairs, creating the hairpin formations that separate sections of the code from the rest of the genome.
Click to read photo caption. By Kevin Weeks, Joseph Watts, and Jason Smith
“I was shocked that we found so much structure,” Watts says. “I thought we’d see a few islands of structure in a mostly unstructured genome.” They compared their model with an analysis done by Kristen Dang, a grad student in biomedical engineering. She used genetic variation among virus particles to find pieces of the genome that evolve slowly because they’re in stable base pairs. She saw many of the same highly structured areas in the genome that Weeks and Watts had found.
So what do these never-before-seen structures actually do? Researchers will spend years answering that question, Weeks says. But he thinks it’s a fair bet that the virus needs many of the structures to survive, because they show up in every version of the fast-mutating HIV genome. Some could be targets for new HIV therapies, although that’s a long way off: scientists have only started to test drugs that target RNA genomes in the past few years.
Still, the way the genome structures interact with the virus’s host cell could give us new ideas for therapies, says Ron Swanstrom, a virologist and an author of the study. “The virus is completely dependent upon the human cells it’s in,” he says. “If we find that there’s a piece of the cell that does something with an RNA structure, and that piece is critical to the virus but not so critical to the cell, we could target that part of the cell to control the virus.”
Weeks and Watts already have an idea that two of the genome structures may set the pace for building viral proteins. HIV grows inside hijacked human cells, using their protein-making machinery to build more of the virus. A cell’s ribosomes read RNA and translate it into chains of amino acids. If these chains come out of the ribosome too quickly, they can interfere with each other as they’re folding into three-dimensional protein shapes. Structures in the HIV genome seem to act like speed bumps, Weeks says, slowing the progress of a ribosome as it reads the RNA sequence.
“We think of it as another level of the genetic code,” Swanstrom says. “You’re encoding protein structure, and it’s a level of complexity that we don’t know much about yet.”
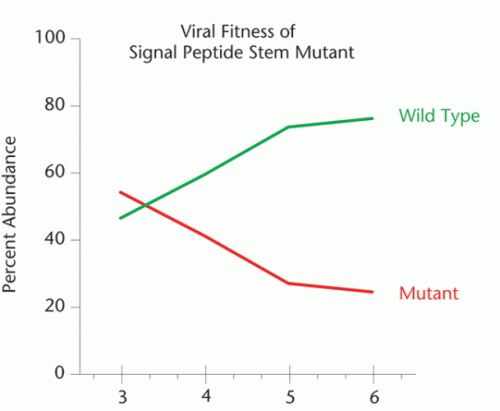
Chart by Elizabeth Pollom. ©2010 Endeavors magazine.
When Pollom mutated an RNA structure in HIV’s Env protein, the resulting mutant virus did not grow as well in cells as the wild type virus.

Click to read photo caption. Chart by Elizabeth Pollom. ©2010 Endeavors magazine.
In Swanstrom’s lab, grad student Elizabeth Pollom is testing the speed-bump hypothesis. Her early results show that when two structures Weeks and Watts found are mutated, the virus doesn’t grow as quickly. Next the lab will try to find out whether messy protein folding is what makes the virus grow slower.
Weeks’s lab is still working on its structure-modeling technology. He wants to make it more sensitive: this first HIV study needed trillions of virus particles, and that’s not the kind of thing you can send away for whenever you want it. “The National Cancer Institute is probably the only place in the world that can hand out enough virus to do a study like this one,” Swanstrom says.
And Weeks does want to do more studies. The published model is a snapshot of the HIV-1 genome at one stage of its complicated replication cycle. “We think there are a lot more structures to find as it goes through all the steps of the cycle,” he says. He wants to decode the HIV-1 genome structure at different stages to make a movie of how it looks over its replication cycle. Like his ambition to decode a whole RNA genome not so long ago, this idea isn’t quite practical yet: it’ll take who-knows-how-many virus particles and whole-genome snapshots to complete. “But in my head,” Weeks says, “I’ve got that next five-year plan.”